Proteomic Laboratory
Research
Requirements for sample preparation
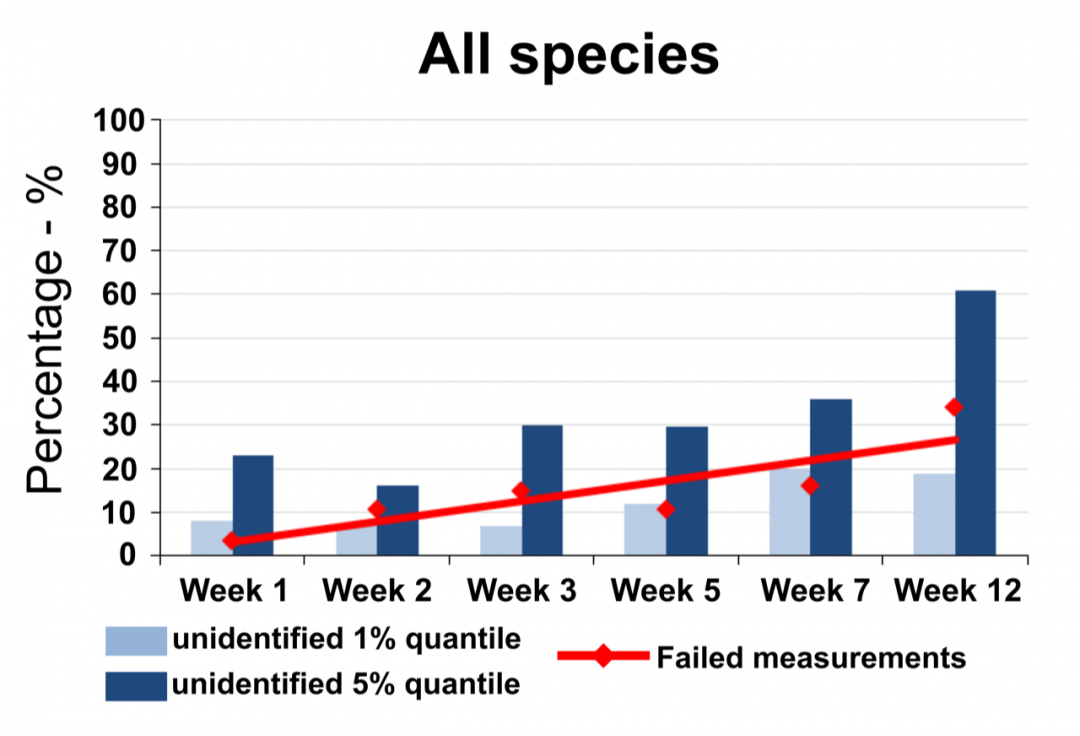
Not all samples can be analyzed using MALDI-TOF mass spectrometry. Certain fixatives and storage conditions can impede successful measurements of the proteome fingerprint for species identification. Hence, one important part of our work is to investigate the storage and fixation of samples to allow species identification using mass spectrometry data. For this, we have carried out extensive studies on meiofauna from stored sediment samples. The investigation of samples fixed in different concentrations of ethanol stored at different temperature conditions revealed that storage temperature influenced mass spectra quality the most. Measurements from samples fixed in ethanol, stored at -25°C showed only minor decrease of signal quality compared to fresh material. In contrast to that, storage at room temperature led to a strong decrease of signal quality within only a few weeks. In concordance with this, also the probability to measure a signal at all decreased with time when samples were stored at room temperature.
Pilot studies for different taxa
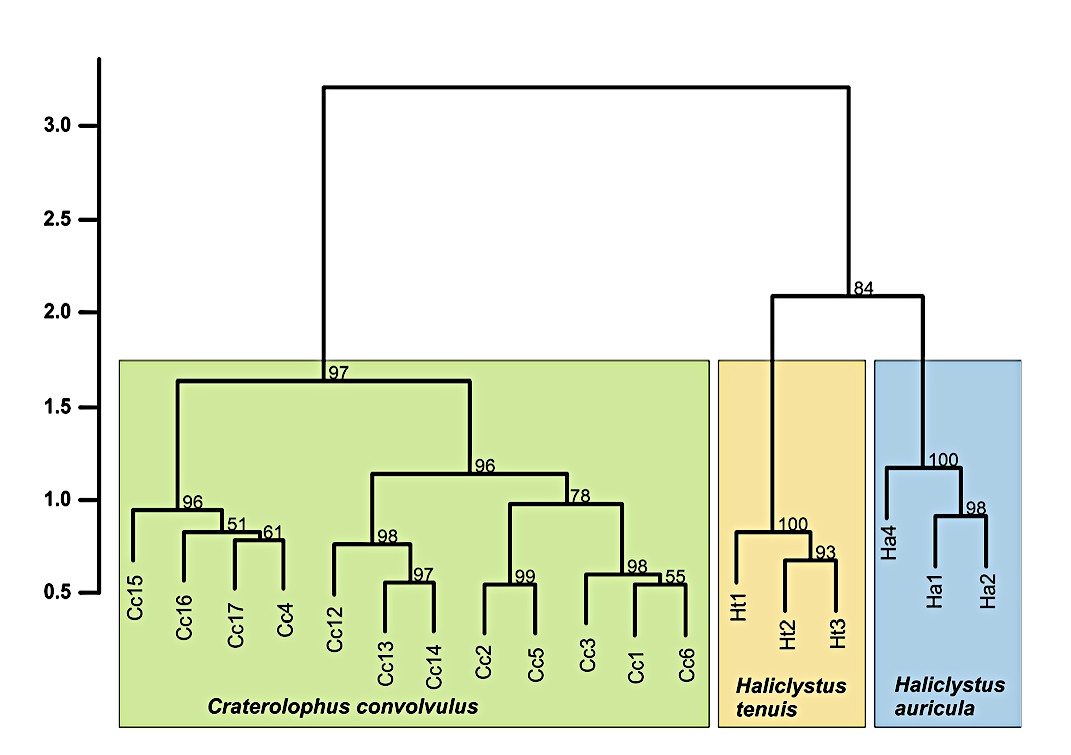
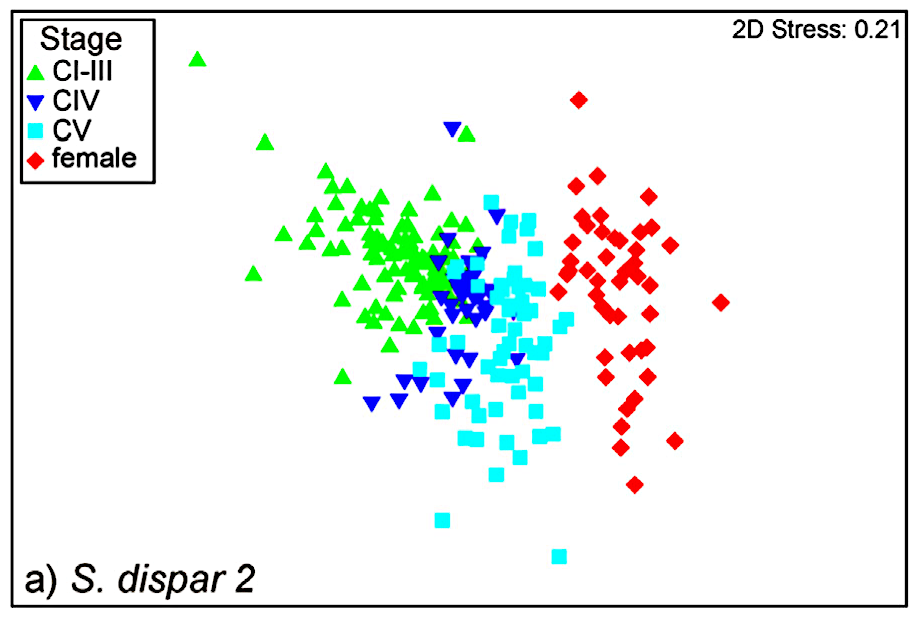
Because the MALDI-TOF MS for identification of metazoan species is not widely applied yet, we are carrying out pilot studies to test if this method allows species identification for a variety of taxa. Animals will be identified both morphologically and with genetic methods such as DNA barcoding before measurements are carried out. If possible, different ontogenetic stages are measured to test if different stages affect the resulting mass spectra. In our lab we are investigating mainly marine animals. For instance, we were able to show that species identification using the proteome fingerprint works good for calanoid copepods from the plankton of the North Sea. For these, even larval stage differentiations were possible. But also for morphological difficult to differentiate cnidarian species, application of MALDI-TOF MS was successful.
Due to previous projects at the DZMB (working group MOLTAX), we have access to a large tissue data base from morphologically and genetically identified species from the North Sea, that will be used in the future for analyses using MALDI-TOF MS.
Methods for rapid biodiversity assessments
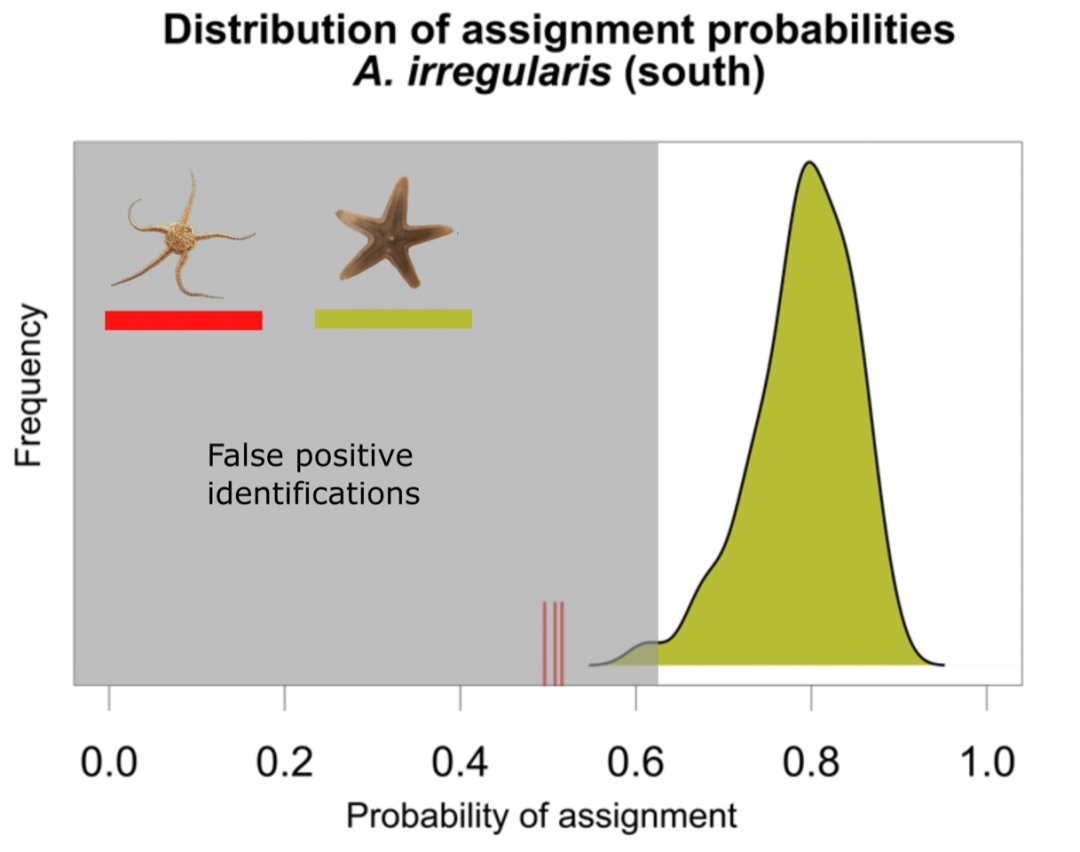
Important for the future application of this method is the availability of data analysis methods. Here, it is important to allow species identification using a reference library. We are working to provide openly accessible software solutions for mass spectrometry data analysis. Using Random Forest, a machine learning tool, we developed a method for species identification including a post hoc test for false positive discovery to detect species that were not part of the applied reference library. In the future, automatic calculation of biodiversity indices based on MALDI-TOF MS data will be part of our work to allow analyzing sampling sites without prior knowledge of the occurring species.
Biodiversity assessments carried out so far
Aside from pilot studies and the development of methods, we have also already carried out biodiversity assessments. Here, we measured the proteome fingerprint for a large number of specimens to assess the species diversity and community structure. First studies showed that communities assessed using MALDI-TOF MS are highly congruent with communities assessed morphologically or based on genetic analyses. Due to this, the time taken to assess community structures could be immensely. Hence, it was possible for deep-sea spinocalanoid copepods to show that vertical habitat preferences play an important role for reproductive isolation of species and coexistence of cryptic species.
In future projects, the method will be used for investigation of the specimen rich meiofauna communities of the North Sea. Also, in the CCZ, a potential area for deep sea mining in the North Pacific, the meiofauna communities will be investigated using the proteome fingerprint. By applying MALDI-TOF MS we expect to accelerate data assessment that was previously done mainly morphologically and hence to be able to focus on new working areas.