Organisms in extreme marine environments: Understanding environmental adaptation and global change effects.
The focus is on mechanisms underlying the colonization of low salinity marine environments. Based on an integrative approach (e.g., population genetics, molecular biology, ecophysiology) we investigate Baltic Mytilus species (representing up to 90% of the benthic biomass) that colonize the Skagerrak/Kattegat (M. edulis; 25-12 practical salinity units (psu)) and the Baltic proper (M. trossulus; 8-4.5 psu). The discovery of massive hybridization (hybrid swarm formation) between both Baltic species led to the hypothesis, that interspecies gene flow may be an essential precondition for the adaptation to extreme salinity conditions. We are testing this hypothesis alongside with alternatives such as demographic effects (larval drift, hybrid zone movement), pre- and postzygotic reproductive traits, differential selection (local environmental adaptation), adaptation, or neutral genetic effects. Apart from a better understanding of evolution, our research contributes to predict consequences of global change. In fact, decreasing salinity levels are predicted for the Baltic which may have consequences on biodiversity of an environment in the neighbourhood of 9 countries and approx. 85 million inhabitants.

Figure 2: Integrative research (genetics, genomics, breeding, ecophysiology, phenotyping) to associate differences in the genomes of Baltic Mytilus species to their ability to colonize the brackish Baltic Sea, i.e., conditions where biomineralization is a physiological challenge.
Key publications
- Knöbel L, Nascimento-Schulze JC, Sanders T, Zeus D, Hiebenthal C, Barboza FR, Stuckas H*, Melzner F* (2021): Salinity driven selection and local adaptation in Baltic Sea Mytilid mussels. Frontiers in Marine Science, published open access on 13th of August 2021. (*shared senior authorship)
- Knoebel L, Breusing C, Bayer T, Sharma V, Hiller M, Melzner F, Stuckas H (2020): Comparative de novo assembly and annotation of mantle transcriptomes from the Mytilus edulis species complex (M. edulis, M. galloprovincialis, M. trossulus). Marine Genomics, 51: Article 100700.
- Stuckas H, Knöbel L, Schade H, Breusing C, Hinrichsen HH, Bartel M., Langguth C, Melzner F (2017): Combining hydrodynamic modelling with genetics: Can passive larval drift shape the genetic structure of Baltic Mytilus populations? Molecular Ecology, 26: 2765-2782.
Biogeography and genetic connectivity in deep sea invertebrates.
While deep sea (water depth below 200m) represents 88% of the ocean’s surface, this part of the earth is largely undiscovered. There is a tremendous lack of knowledge about aspects such as biodiversity or ecosystem function. Consequently, we use molecular approaches (e.g., molecular taxonomy, population genetics) to discover species, their distribution and biogeography as well as issues related to genetic diversity, divergence and connectivity. The applied aspect of this research refers to impact assessments related to planned deep sea mining along ocean ridges (inactive hydrothermal vents) or abyssal plains (manganese nodules).
Key publications
- Janssen A*, Stuckas H*, Annemiek Wink, Pedro Martinez Arbizu P (2019): Patterns of genetic connectivity and demograhy in populations of predominant macrofaunal taxa (Polychaeta, Isopoda) in abyssal polymetallic nodule fields: Implications for conservation management. Marine Biodiversity, 49: 2641–2658. (*shared first authorship)
- Dunn DC, Van Dover CL, Etter RJ, Smith CR, Levin LA, T. Morato, Colaço A, Dale AC, Gebruk AV, Gjerde KM, Halpin PN, Howell KL, Johnson D, Perez JAA, Ribeiro MC, Stuckas H, P. Weaver and the SEMPIA Workshop Participants (2018): A strategy for the conservation of biodiversity on mid-ocean ridges from deep-sea mining. Science Advances, 4(2): eaar4313.
- Gollner G*, Stuckas H*, Kihara TC, Kodami S, Martinez Arbizu P (2016): Mitochondrial DNA analyses indicate high diversity, expansive population growth and high genetic connectivity of vent copepods (Dirivultidae) across different oceans. Plos One, 11(10): e0163776. (* shared co-authorship)
Spatial genetic structure of populations and hybridization
This research focus is concerned with assessing the spatial genetic structure of populations. It includes genetic monitoring of endangered species based on minimal invasive sampling (e.g., COI-barcoding of mammals using scat). Apart from contributing to conservation issues, we aim at identifying cases of interspecific gene flow as successfully done in various birds and turtles/terrapins. Together with our research on the hybridizing Baltic mussels (see above), these activities aim at exploring the biological significance of hybridization (e.g., speciation, amalgamation of gene pools, speciation in reverse).



Figure 3: The population genetics group collaborates with other scientists and perfomes reserac ins the fields of biomonitoring, speciation, hybridization, and conservation on various organisms.
Key publications
- Wolfgramm H, Martens J, Töpfer T, Vamberger M, Pathak A, Stuckas H, Päckert M (2021): Asymmetric allelic introgression across a hybrid zone of the coal tit (Periparus ater) in the central Himalayas. Ecology and Evolution, 11: 17332-17351.
- Päckert M, Bekacem, AA, Wolfgramm H, Gast O, Canal D, Giacalone G, Lo Valvo M, Vamberger M, Wink M, Martens J, Stuckas H (2019): Genetic admixture despite ecological segregation in a North African sparrow hybrid zone (Aves, Passeriformes, Passer domesticus x Passer hispaniolensis). Ecology and Evolution, 9: 12710 – 12726.
- Vamberger M. Stuckas H, Vargas-Ramírez M, Kehlmaier C, Ayaz D, Aloufi AA, Lymberakis P, Široký P, Fritz U (2017): Unexpected hybridization patterns in Near Eastern terrapins (Mauremys caspica, M. rivulata) indicate ancient gene flow across the Fertile Crescent. Zoologica Scripta, 46: 401-413.
Linking phenotypic differences between species to differences in their genomes using Forward Genomics.
Discovering genomic causes of phenotypes is one major challenge in bioscience that can be tackled based on advances in ‘OMICS-technologies. While our research on organisms in extreme marine environments integrates phenotype assessment with population genetics (intraspecific level and hybridization between sister species), we co-initiated a long term project that aims at linking phenotypic differences between species with differences in their genomes (evolutionary scale, body plan evolution). We currently focus on mammalian species and explore publically available mammalian genomes using the novel concept of Forward Genomics. This concept basically correlates genome alignments with phenotype matrices to identifying genomic loci that explain the absence of traits in some mammalian species while the same traits are present in others (convergent trait loss). The availability of phenotypic trait information in a format suitable for analyses in Forward Genomics can be considered a strong bottleneck in bioscience! For instance, information must be presented in distinct categories (e.g., present/absent) or in a machine actionable format (e.g., NEXUS file). Thus, one main task to be accomplished was to establish a novel data platform, i.e., MaTrics (Mammalian Traits for comparative genomics). MaTrics will be developed further in the near future (https://www.senckenberg.de/de/institute/senckenberg-naturhistorische-sammlungen-dresden/museum-fuer-tierkunde/dd-sekt-mammalogie/matrics-consortium/). By focussing on anatomical traits (e.g., position of testis) or nutrition we successfully identified gene losses associated to phenotypic differences between mammals. This did we not only deepen our understanding of evolutionary processes but has also relevance to understand human organ function (phylomedicine). Our long-term vision is to apply the conceptual framework for Forward Genomics to non-mammalian taxa. For instance, this may allow understanding genomic changes associated with adaptation to different salinity or temperature conditions (see research focus on salinity adaptation above).
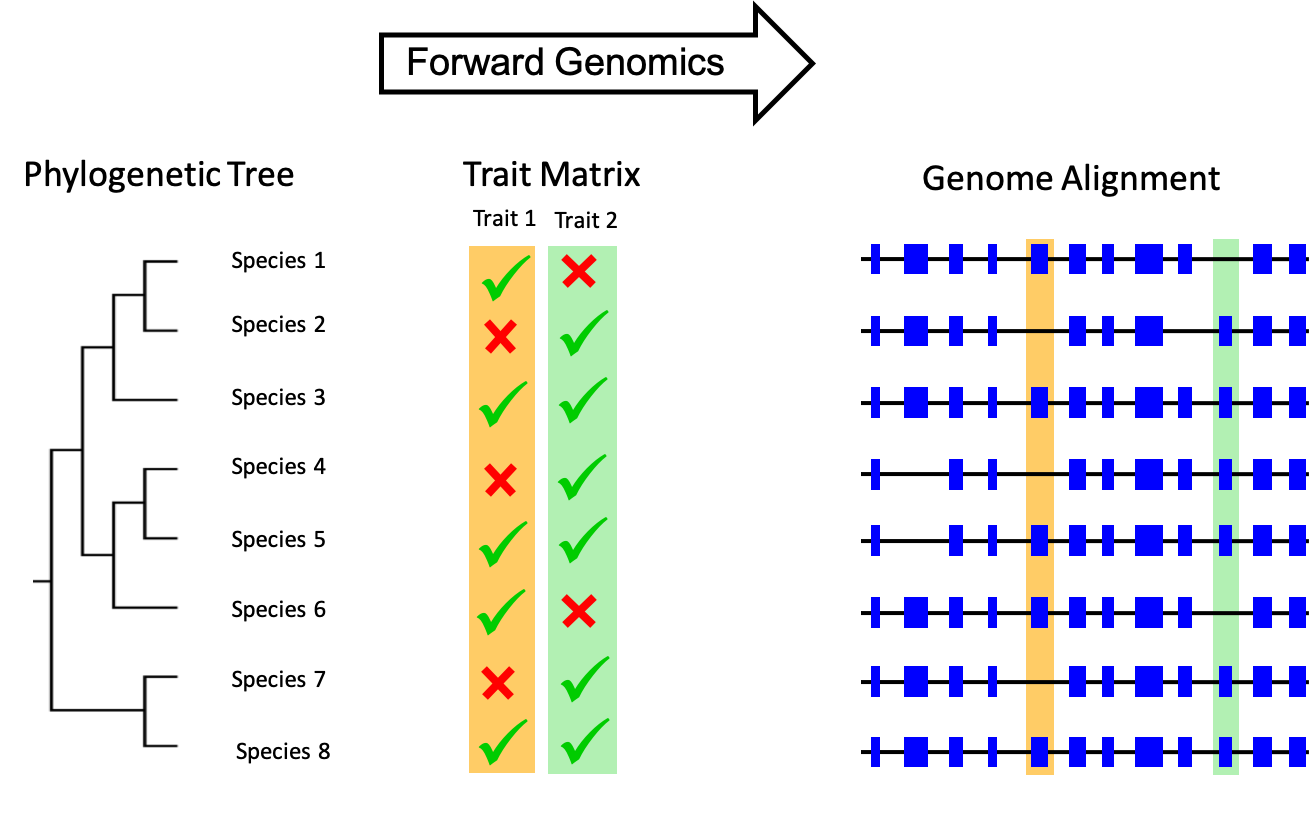

Figure 4: Forward Genomics aims at exploring which genomic differences underlie the phenotypic variability for instance in mammals.
Key publications
- Wagner F, Ruf I, Lehmann T, Hofmann R, Ortmann S, Schiffmann C, Hiller M, Stefen C, Stuckas H (2022): Reconstruction of evolutionary changes in fat and toxin consumption reveals associations with gene losses in mammals: a case study for the lipase inhibitor PNLIPRP1 and the xenobiotic receptor NR1I3. Journal of Evolutionary Biology, 35: 225-239.
- Stefen C, Wagner F, Aszatlos M, Giere P, Grobe P, Hiller M, Hofmann R, Jähde M, Lächele U, Lehmann T, Ortmann S, Peters B, Ruf I, Schiffmann C, Their N, Unterhitzenberger G, Vogt L, Rudolf M, Wehner P, Stuckas H (2021): Phenotyping in the era of genomics: MaTrics – a digital character matrix to document mammalian phenotypic traits. Mammalian Biology, published open access on 7th of December 2021.
- Sharma V, Lehmann T, Stuckas H, Hiller M (2018): Loss of the gubernaculum-inducing INSL3 and RXFP2 genes in afrotheria shows that testicular descent is the ancestral condition in placental mammals. Plos Biology, 16(6): e2005293.